Test: Gas Evolution
In this chapter we are comparing the gas evolution to the analytical solution.
Analytical Solution
Viscous gas accretion has analytical solutions as given by Lynden-Bell & Pringle (1974) or Hartmann et al. (1998).
We assume a viscously evolving disk
\(\Large \frac{\partial}{\partial t} \Sigma_\mathrm{g} = \frac{3}{r}\frac{\partial}{\partial r} \left[ \sqrt{r} \frac{\partial}{\partial r} \left( \sqrt{r} \nu \Sigma_\mathrm{g} \right) \right]\)
with an \(\large \alpha\)-viscosity prescription given by
\(\Large \begin{split} \nu &= \alpha \frac{c_\mathrm{s}^2}{\Omega_\mathrm{K}}\\ &\propto r^{s} \cdot r^{q} \cdot r^{\frac{3}{2}} = r^\gamma. \end{split}\)
In the default simulation we have \(\large s=0\), \(\large q=-\frac{1}{2}\), and therefore \(\large \gamma=1\).
This differential equation for viscous accretion has self-similar solutions given by
\(\Large \Sigma_\mathrm{g} = \frac{C}{3\pi\nu_1R^\gamma}T^{-\frac{5/2-\gamma}{2-\gamma}} \exp \left[ -\frac{R^{2-\gamma}}{T} \right]\)
with a dimensionless scaling of the grid \(\large R=\frac{r}{r_1}\), the radial scaling factor \(\large r_1\) and the viscosity at the scaling location \(\large \nu_1=\nu\left(r_1\right)\).
\(\Large T=\frac{t}{t_\mathrm{s}}+1\) is the dimensionless time and
\(\Large t_\mathrm{s} = \frac{1}{3 \left( 2-\gamma \right)^2}\frac{r_1^2}{\nu_1}\).
\(\large C\) is a scaling constant that defines the initial mass of the disk \(\large M_0\)
\(\Large C = M_0 \frac{3\nu_1}{2r_1^2\left(2-\gamma\right)}\).
We can now write a function that returns the surface density profile at any given time to compare it against the simulation result.
[1]:
def LBP74_analytical(r, rc, nu, M0, t):
"""Function calculates the self-similar solution of Lynden-Bell & Pringle (1974)
It is assuming that the viscosity follows a power law.
Parameters
----------
r : Field
Radial grid
rc : float
Initial critical cutoff radius
nu : Field
Kinematic viscosity
M0 : float
Initial disk mass
t : float
Time
Returns
-------
Sigma : Field
Gas surface density profile at time t
"""
import numpy as np
# Get power law index of viscosity
gamma = np.mean( np.diff( np.log10(nu) ) / np.diff( np.log10(r) ) )
# Convert to dimensionless quantities
R = r/rc
nu1 = nu[0] * (rc/r[0])**gamma
ts = rc**2 / (3*nu1*(2-gamma)**2)
T = t/ts + 1
# Calculate scaling constant
C = (2-gamma)*M0 * 3*nu1/(2*rc**2)
return C / (3*np.pi*nu1*R**gamma) * T**(-(5/2-gamma)/(2-gamma)) * np.exp(-R**(2-gamma)/T)
We test the gas evolution for the default model.
[2]:
from dustpy import Simulation
sim = Simulation()
sim.initialize()
[3]:
import matplotlib.pyplot as plt
We want to compare the simulation to the analytical solution at four discrete snapshots between \(10,000\) and \(10,000,000\) years.
[4]:
import dustpy.constants as c
import numpy as np
snapshots = np.hstack(
[sim.t, np.geomspace(1.e4, 1.e7, num=4) * c.year]
)
[5]:
fig = plt.figure(dpi=150)
ax = fig.add_subplot(111)
ax.loglog(sim.grid.r/c.au, sim.gas.Sigma, c="black", lw=3, alpha=0.25, label="DustPy initial")
ax.loglog(sim.grid.r/c.au, LBP74_analytical(sim.grid.r, sim.ini.gas.SigmaRc, sim.gas.nu, sim.ini.gas.Mdisk, 0), "--", lw=1, c="C0", label="t = 0.0e+00 yrs")
for i in range(1, len(snapshots)):
cstr = "C" + str(i)
ax.loglog(sim.grid.r/c.au, LBP74_analytical(sim.grid.r, sim.ini.gas.SigmaRc, sim.gas.nu, sim.ini.gas.Mdisk, snapshots[i]), "--", lw=1, c=cstr, label="t = {:3.1e} yrs".format(snapshots[i]/c.year))
ax.legend()
ax.set_xlim(sim.grid.r[0]/c.au, sim.grid.r[-1]/c.au)
ax.set_ylim(1.e-2, 1.e4)
ax.set_xlabel("Distance from star [au]")
ax.set_ylabel("Surface density [g/cm²]")
fig.tight_layout()
plt.show()
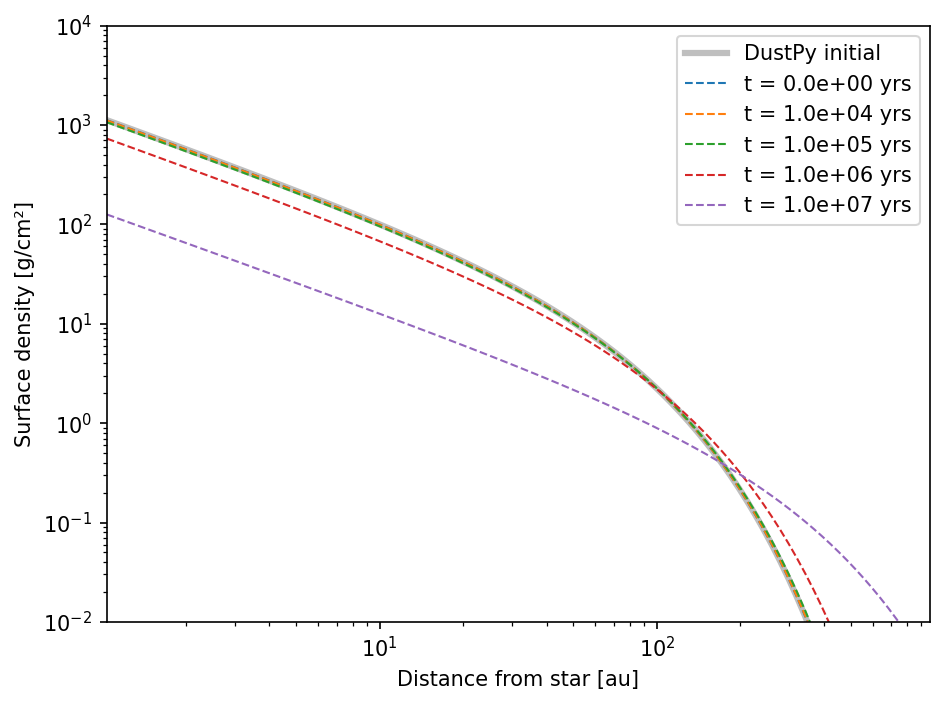
Setting up DustPy
We now set up a DustPy simulation to compare the gas evolution to the analytical solutions.
Turning off Dust Evolution
Since we’re not interested in the dust evolution and since this is the computationally most expensive part of DustPy, we simply turn it off. We do this by removing the integration instruction and unsetting all updater that are not needed.
[6]:
sim.dust.S.coag[...] = 0.
sim.dust.S.coag.updater = None
sim.dust.S.ext[...] = 0.
sim.dust.S.ext.updater = None
sim.dust.S.hyd[...] = 0.
sim.dust.S.hyd.updater = None
sim.dust.S.tot[...] = 0.
sim.dust.S.tot.updater = None
sim.dust.S.updater = None
sim.update()
[7]:
sim.integrator.instructions
[7]:
[Instruction (Dust: implicit 1st-order direct solver),
Instruction (Gas: implicit 1st-order direct solver)]
[8]:
del(sim.integrator.instructions[0])
[9]:
sim.integrator.instructions
[9]:
[Instruction (Gas: implicit 1st-order direct solver)]
Modifying Integration Variable
We set the desired snapshots, that we already defined above.
[10]:
sim.t.snapshots = snapshots
Setting Writer Options
We also change the name of the output directory and allow DustPy to overwrite existing data files.
[11]:
sim.writer.datadir = "test_gas_evolution"
sim.writer.overwrite = True
We can now run the simulation.
[12]:
sim.run()
DustPy v1.0.8
Documentation: https://stammler.github.io/dustpy/
PyPI: https://pypi.org/project/dustpy/
GitHub: https://github.com/stammler/dustpy/
Please cite Stammler & Birnstiel (2022).
Checking for mass conservation...
- Sticking:
max. rel. error: 2.81e-14
for particle collision
m[114] = 1.93e+04 g with
m[116] = 3.73e+04 g
- Full fragmentation:
max. rel. error: 6.66e-16
for particle collision
m[90] = 7.20e+00 g with
m[95] = 3.73e+01 g
- Erosion:
max. rel. error: 1.78e-15
for particle collision
m[110] = 5.18e+03 g with
m[118] = 7.20e+04 g
Creating data directory test_gas_evolution.
Writing file test_gas_evolution/data0000.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0001.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0002.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0003.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0004.hdf5
Writing dump file test_gas_evolution/frame.dmp
Execution time: 0:00:08
Reading and Plotting Data
We now read the data and compare it to the analytical solutions.
[13]:
r = sim.writer.read.sequence("grid.r") / c.au
Sigma = sim.writer.read.sequence("gas.Sigma")
nu = sim.writer.read.sequence("gas.nu")
t = sim.writer.read.sequence("t") / c.year
[14]:
fig = plt.figure(dpi=150)
ax = fig.add_subplot(111)
ax.loglog(r[0], Sigma[0], c="C0", label="t = 0.0e+00 yrs")
for i in range(1, len(sim.t.snapshots)):
cstr = "C"+str(i)
ax.loglog(r[i], LBP74_analytical(r[i]*c.au, sim.ini.gas.SigmaRc, nu[0], sim.ini.gas.Mdisk, t[i]*c.year), "--", lw=1, c=cstr)
ax.loglog(r[i], Sigma[i], c=cstr, label="t = {:3.1e} yrs".format(t[i]))
ax.plot(0., 0., ":", c="black", label="analytical")
ax.legend()
ax.set_xlim(r[0, 0], r[0, -1])
ax.set_ylim(1.e-2, 1.e4)
ax.set_xlabel("Distance from star [au]")
ax.set_ylabel("Surface density [g/cm²]")
fig.tight_layout()
plt.show()
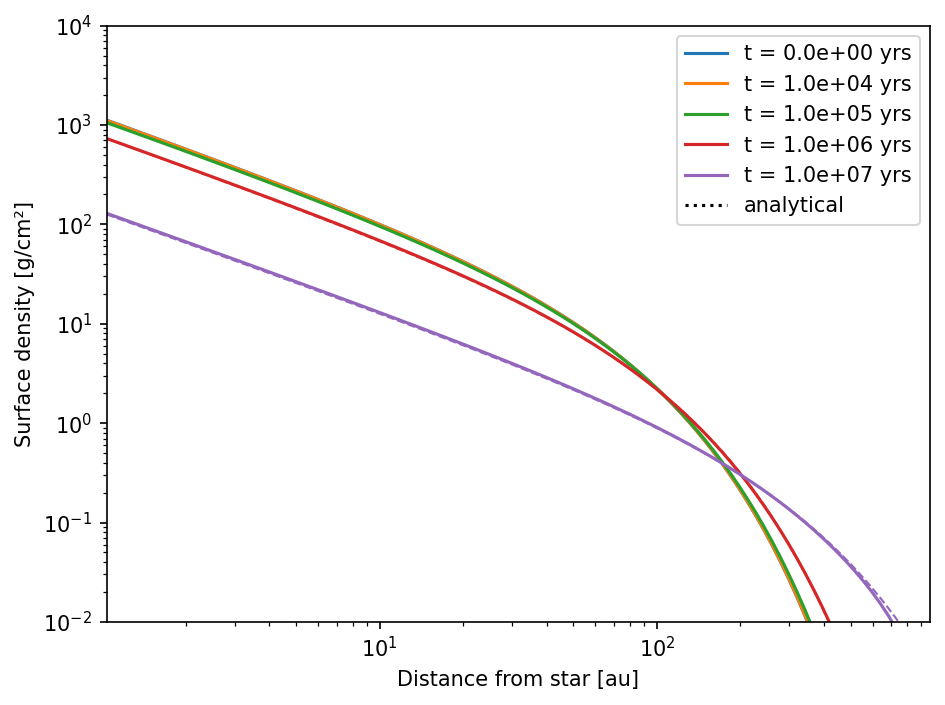
As you can see, the time evolution of the gas disk (in the default model) is in good agreement with the analytical solutions.
Modifying the Power Law Index
We now run the same test again, but we change the initial power law index of the gas surface density profile from \(-1.0\) to \(-1.5\).
[15]:
sim = Simulation()
sim.ini.gas.SigmaExp = -1.5
[16]:
sim.initialize()
[17]:
sim.dust.S.coag[...] = 0.
sim.dust.S.coag.updater = None
sim.dust.S.ext[...] = 0.
sim.dust.S.ext.updater = None
sim.dust.S.hyd[...] = 0.
sim.dust.S.hyd.updater = None
sim.dust.S.tot[...] = 0.
sim.dust.S.tot.updater = None
sim.dust.S.updater = None
sim.update()
[18]:
del(sim.integrator.instructions[0])
[19]:
sim.t.snapshots = snapshots
[20]:
sim.writer.datadir = "test_gas_evolution"
sim.writer.overwrite = True
[21]:
sim.run()
DustPy v1.0.8
Documentation: https://stammler.github.io/dustpy/
PyPI: https://pypi.org/project/dustpy/
GitHub: https://github.com/stammler/dustpy/
Please cite Stammler & Birnstiel (2022).
Checking for mass conservation...
- Sticking:
max. rel. error: 2.81e-14
for particle collision
m[114] = 1.93e+04 g with
m[116] = 3.73e+04 g
- Full fragmentation:
max. rel. error: 6.66e-16
for particle collision
m[90] = 7.20e+00 g with
m[95] = 3.73e+01 g
- Erosion:
max. rel. error: 1.78e-15
for particle collision
m[110] = 5.18e+03 g with
m[118] = 7.20e+04 g
Writing file test_gas_evolution/data0000.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0001.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0002.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0003.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0004.hdf5
Writing dump file test_gas_evolution/frame.dmp
Execution time: 0:00:11
[22]:
r = sim.writer.read.sequence("grid.r") / c.au
Sigma = sim.writer.read.sequence("gas.Sigma")
nu = sim.writer.read.sequence("gas.nu")
t = sim.writer.read.sequence("t") / c.year
[23]:
fig = plt.figure(dpi=150)
ax = fig.add_subplot(111)
ax.loglog(r[0], Sigma[0], c="C0", label="t = 0.0e+00 yrs")
for i in range(1, len(sim.t.snapshots)):
cstr = "C"+str(i)
ax.loglog(r[i], LBP74_analytical(r[i]*c.au, sim.ini.gas.SigmaRc, nu[0], sim.ini.gas.Mdisk, t[i]*c.year), "--", lw=1, c=cstr)
ax.loglog(r[i], Sigma[i], c=cstr, label="t = {:3.1e} yrs".format(t[i]))
ax.plot(0., 0., ":", c="black", label="analytical")
ax.legend()
ax.set_xlim(r[0, 0], r[0, -1])
ax.set_ylim(1.e-2, 1.e4)
ax.set_xlabel("Distance from star [au]")
ax.set_ylabel("Surface density [g/cm²]")
fig.tight_layout()
plt.show()
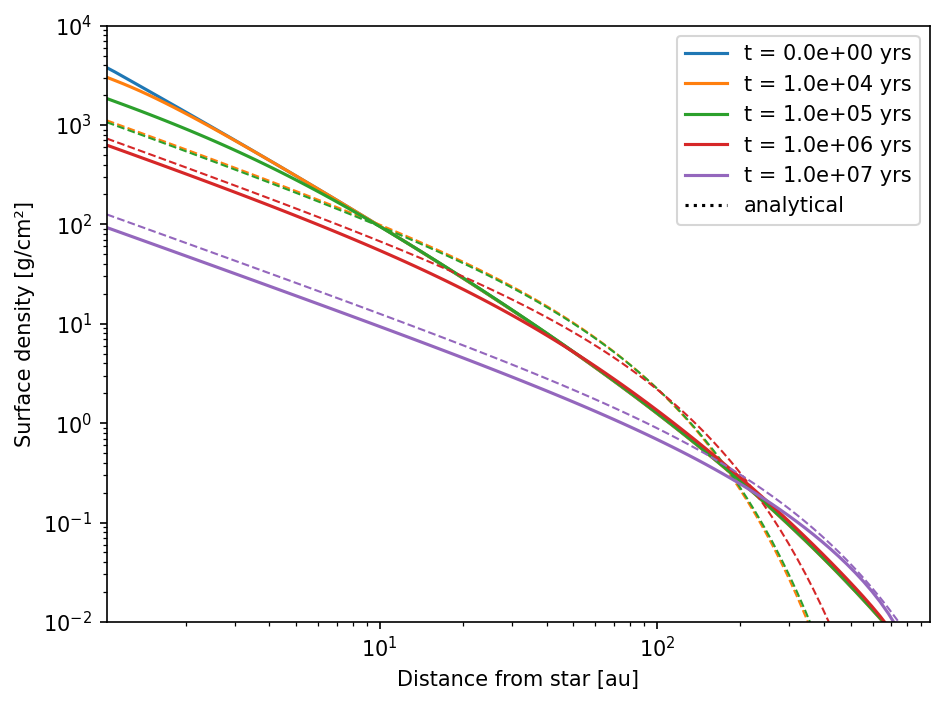
\(\Large \frac{\partial}{\partial r} \left( 3\pi\Sigma_\mathrm{g}\nu \right) = \text{const.}\)
Or in other words
\(\Large \Sigma_\mathrm{g}\nu \propto r^p r^\gamma = r^0 \\ \Rightarrow p+\gamma = 0\)
We’ve change \(\large p\) from \(-1\) to \(-1.5\), but we have not changed \(\large \gamma\), which is still \(1\) from the default setup as described above. Therefore, in our initial conditions we have
\(\Large p+\gamma=-0.5 \neq 0\).
Over time the system finds a steady state. Since the temperature profile and the \(\large \alpha\)-viscosity parameter are both fix in the default model, the only way for the system to get into steady state is by changing the slope of the gas surface density. This is what happened here: the surface density profile went from \(-1.5\) initially to \(-1\) after \(10\,\)Myrs.
To fix this problem we need to change the viscosity of the setup. We can do this by either changing the temperature profile, or by changing the \(\large \alpha\)-profile. We are changing \(\large \alpha\) here
\(\Large \alpha \left( r \right) \propto r^\frac{1}{2}\)
which leads to \(\large \gamma = 1.5\).
[24]:
sim = Simulation()
sim.ini.gas.SigmaExp = -1.5
sim.initialize()
[25]:
sim.gas.alpha[:] = sim.ini.gas.alpha * (sim.grid.r[:]/sim.ini.gas.SigmaRc)**0.5
[26]:
sim.dust.S.coag[...] = 0.
sim.dust.S.coag.updater = None
sim.dust.S.ext[...] = 0.
sim.dust.S.ext.updater = None
sim.dust.S.hyd[...] = 0.
sim.dust.S.hyd.updater = None
sim.dust.S.tot[...] = 0.
sim.dust.S.tot.updater = None
sim.dust.S.updater = None
sim.update()
[27]:
del(sim.integrator.instructions[0])
[28]:
sim.t.snapshots = snapshots
[29]:
sim.writer.datadir = "test_gas_evolution"
sim.writer.overwrite = True
[30]:
sim.run()
DustPy v1.0.8
Documentation: https://stammler.github.io/dustpy/
PyPI: https://pypi.org/project/dustpy/
GitHub: https://github.com/stammler/dustpy/
Please cite Stammler & Birnstiel (2022).
Checking for mass conservation...
- Sticking:
max. rel. error: 2.81e-14
for particle collision
m[114] = 1.93e+04 g with
m[116] = 3.73e+04 g
- Full fragmentation:
max. rel. error: 6.66e-16
for particle collision
m[90] = 7.20e+00 g with
m[95] = 3.73e+01 g
- Erosion:
max. rel. error: 1.78e-15
for particle collision
m[110] = 5.18e+03 g with
m[118] = 7.20e+04 g
Writing file test_gas_evolution/data0000.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0001.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0002.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0003.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0004.hdf5
Writing dump file test_gas_evolution/frame.dmp
Execution time: 0:00:20
[31]:
r = sim.writer.read.sequence("grid.r") / c.au
Sigma = sim.writer.read.sequence("gas.Sigma")
nu = sim.writer.read.sequence("gas.nu")
t = sim.writer.read.sequence("t") / c.year
[32]:
fig = plt.figure(dpi=150)
ax = fig.add_subplot(111)
ax.loglog(r[0], Sigma[0], c="C0", label="t = 0.0e+00 yrs")
for i in range(1, len(sim.t.snapshots)):
cstr = "C"+str(i)
ax.loglog(r[i], LBP74_analytical(r[i]*c.au, sim.ini.gas.SigmaRc, nu[0], sim.ini.gas.Mdisk, t[i]*c.year), "--", lw=1, c=cstr)
ax.loglog(r[i], Sigma[i], c=cstr, label="t = {:3.1e} yrs".format(t[i]))
ax.plot(0., 0., ":", c="black", label="analytical")
ax.legend()
ax.set_xlim(r[0, 0], r[0, -1])
ax.set_ylim(1.e-2, 1.e4)
ax.set_xlabel("Distance from star [au]")
ax.set_ylabel("Surface density [g/cm²]")
fig.tight_layout()
plt.show()
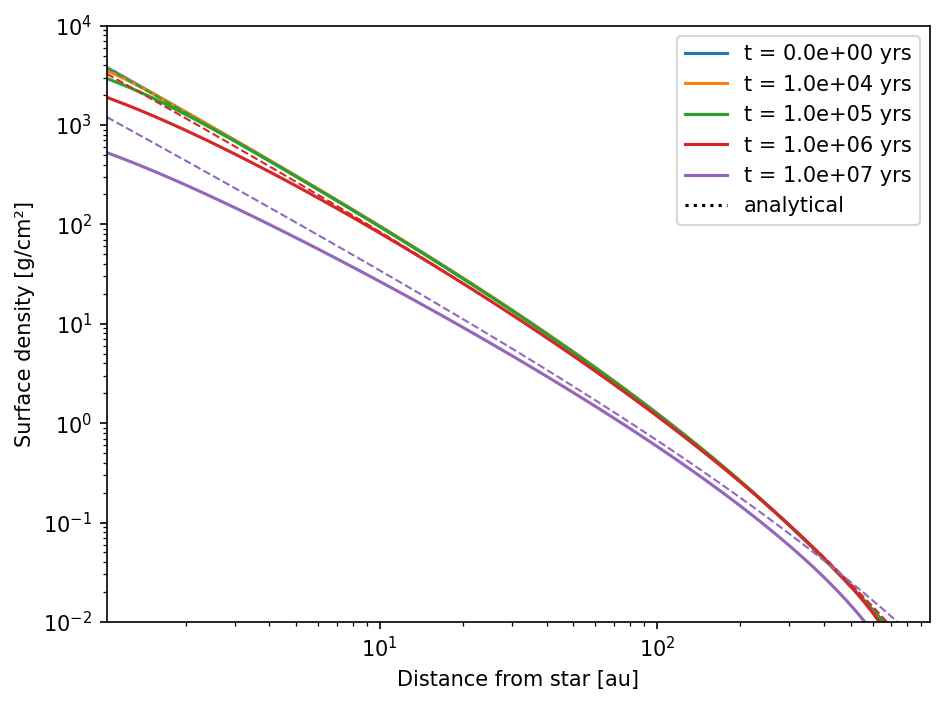
This is closer to the analytical solutions. However, there are stil deviations at the inner boundary of the grid.
\(\Large \frac{1}{r}\frac{\partial}{\partial r} \left( r\Sigma_\mathrm{g} \right) = \text{const.}\)
This can be set exactly within a linear system, such as the one that is solved for gas evolution. A surface density profile of \(-1\) as is set for the default model has automatically a constant gradient at the inner boundary. But a surface density profile of \(-1.5\) does not. Here we need to change the inner boundary condition to constant power law.
[33]:
sim = Simulation()
sim.ini.gas.SigmaExp = -1.5
sim.initialize()
[34]:
sim.gas.alpha[:] = sim.ini.gas.alpha * (sim.grid.r[:]/sim.ini.gas.SigmaRc)**0.5
[35]:
sim.dust.S.coag[...] = 0.
sim.dust.S.coag.updater = None
sim.dust.S.ext[...] = 0.
sim.dust.S.ext.updater = None
sim.dust.S.hyd[...] = 0.
sim.dust.S.hyd.updater = None
sim.dust.S.tot[...] = 0.
sim.dust.S.tot.updater = None
sim.dust.S.updater = None
sim.update()
[36]:
del(sim.integrator.instructions[0])
[37]:
sim.gas.boundary.inner.setcondition("const_pow")
[38]:
sim.t.snapshots = snapshots
[39]:
sim.writer.datadir = "test_gas_evolution"
sim.writer.overwrite = True
[40]:
sim.run()
DustPy v1.0.8
Documentation: https://stammler.github.io/dustpy/
PyPI: https://pypi.org/project/dustpy/
GitHub: https://github.com/stammler/dustpy/
Please cite Stammler & Birnstiel (2022).
Checking for mass conservation...
- Sticking:
max. rel. error: 2.81e-14
for particle collision
m[114] = 1.93e+04 g with
m[116] = 3.73e+04 g
- Full fragmentation:
max. rel. error: 6.66e-16
for particle collision
m[90] = 7.20e+00 g with
m[95] = 3.73e+01 g
- Erosion:
max. rel. error: 1.78e-15
for particle collision
m[110] = 5.18e+03 g with
m[118] = 7.20e+04 g
Writing file test_gas_evolution/data0000.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0001.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0002.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0003.hdf5
Writing dump file test_gas_evolution/frame.dmp
Writing file test_gas_evolution/data0004.hdf5
Writing dump file test_gas_evolution/frame.dmp
Execution time: 0:00:19
[41]:
r = sim.writer.read.sequence("grid.r") / c.au
Sigma = sim.writer.read.sequence("gas.Sigma")
nu = sim.writer.read.sequence("gas.nu")
t = sim.writer.read.sequence("t") / c.year
[42]:
fig = plt.figure(dpi=150)
ax = fig.add_subplot(111)
ax.loglog(r[0], Sigma[0], c="C0", label="t = 0.0e+00 yrs")
for i in range(1, len(sim.t.snapshots)):
cstr = "C"+str(i)
ax.loglog(r[i], LBP74_analytical(r[i]*c.au, sim.ini.gas.SigmaRc, nu[0], sim.ini.gas.Mdisk, t[i]*c.year), "--", lw=1, c=cstr)
ax.loglog(r[i], Sigma[i], c=cstr, label="t = {:3.1e} yrs".format(t[i]))
ax.plot(0., 0., ":", c="black", label="analytical")
ax.legend()
ax.set_xlim(r[0, 0], r[0, -1])
ax.set_ylim(1.e-2, 1.e4)
ax.set_xlabel("Distance from star [au]")
ax.set_ylabel("Surface density [g/cm²]")
fig.tight_layout()
plt.show()
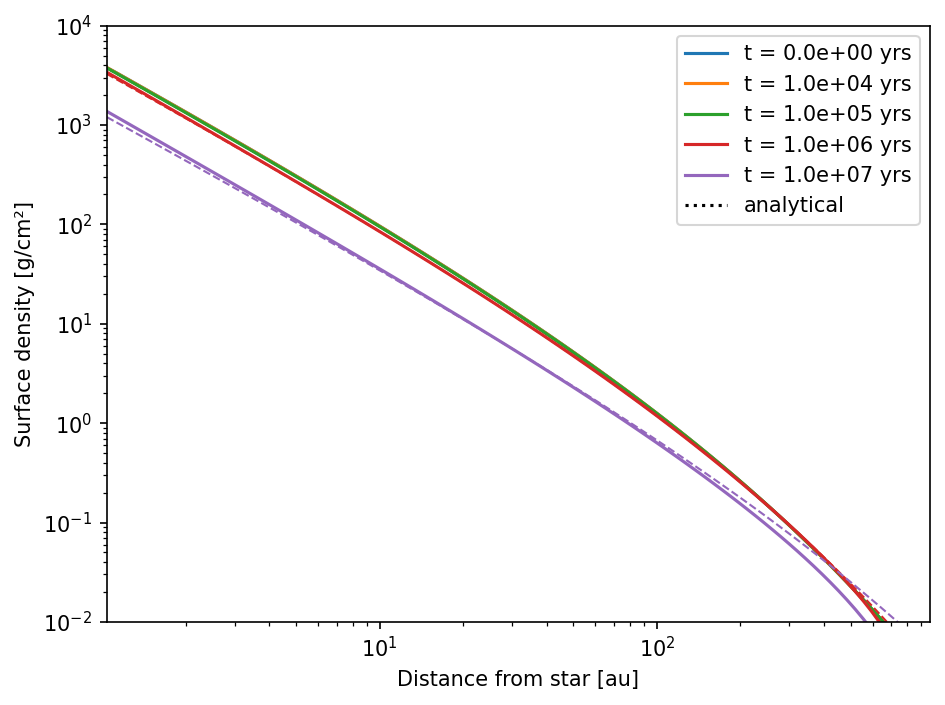
This is not perfect, but as good as it can get. The deviation at the outer boundary is due to the fact that the outer boundary conditions is setting the gas to its floor value to prevent inflow.
In general: Always test your models against analytical solutions, if possible and decide then if the errors are acceptable for you.
The reason for that is that a constant power law is not necessarily possible within a linear system. A power of \(-1.5\) for example is not.
DustPy is “cheating” here a bit. A boundary condition of const_pow is behind the scenes actually a boundary condition of val, where the value is calculated from the current surface density. This is close enough to the actual value in most cases. After the integration the boundary condition is then strictly enforced, by setting the correct power with the new values of the surface density.
Since the default model uses a power of \(-1\) and since this can be done exactly with the const_grad boundary condition, we prefer it over the const_pow condition in the default setup.